Technology and Innovation
Improvement of Fundamental Technologies for Post-translational Modification Proteomics
In this project, techniques for sample extraction and preparation, mass spectrometric identification of post-translational modification (PTM), molecular imaging, etc. are invest igated to establish fundamental technologies for clinical proteomics of PTM.
Techniques have been established 1) to detect and identify ~3,000 plasma proteins and >2,000 phosphorylated proteins (>4,000 phosphorylated peptides) by mass spectrometry (Fig. 4); 2) to analyze quantitatively and qualitatively phosphorylated proteins by affinity electrophoresis; 3) to identify proteins on membrane filters by mass spectrometry; 4) and to perform molecular imaging, etc.
Using these techniques, phosphorylated proteins from culture cells or patient tissues of ovarian cancer, prostate cancer and gastrointestinal stromal tumor were comprehensively detected and identified by mass spectrometry. In this analysis, a total of ~10,000 phosphorylated proteins (~20,000 phosphorylated peptides) were detected for a few weeks. Among them, ~240 disease-associated phosphorylated proteins were identified. The number of phosphorylated proteins detected is beyond our expectations.
On the other hand, recently, techniques using antibodies, siRNA, model animals, two-photon excitation fluorescence microscopy, NMR and X-ray crystallographic analysis have been developed to analyze protein function and the relation between proteins and diseases. In this project, these techniques were improved for analysis of PTM, and using the improved techniques, more effective analysis is performed to determine the relation between PTM and diseases.
In addition, assay techniques to detect PTM proteins as diagnosis biomarkers are developed. The possibility to use abnormal PTM proteins as PET markers and to detect the PTM proteins by selected reaction monitoring of mass spectrometry is investigated.
To Create a Novel Medical Technology
1) A Novel Strategy to Explore Drug Targets and Diagnostic Biomarkers
In the exploration of drug targets and diagnostic biomarkers, diseaseassociated proteins are detected. After verification and validation, they are used as drug targets and diagnostic biomarkers. However, previously, in most cases, disease-associated proteins were detected by chance, and their potential value in the drug development pipeline was verified and validated over a significant period of time. For example, the prostate marker glycoprotein PSA was discovered in 1847, and FDA approved the use of this protein as a biomarker in 1998. In this project, we comprehensively analyze proteins to explore systematically disease-associated proteins. We have detected many promising candidates as drug targets and diagnostic biomarkers. We wish to translate these proteins to practical use as drug targets and diagnostic biomarkers over a short period.
2) Post-translationally Modified Proteins as Drug Targets and Diagnostic Biomarkers
Proteins are post-translationally modified and this post-translational modification process leads proteins to function. However, abnormal post-translational modification of proteins invariably changes the function of the protein, and such changes are often associated with various diseases. There are several reports describing diseases caused by abnormal post-translational modifications. However, we still need to further develop an understanding of the relation between post-translational modifications and diseases.
It is possible to use abnormal post-translationally modified proteins as drug targets and diagnostic biomarkers. In this project, we have been investigating the relation between abnormal post-translational modifications and diseases, and we have identified various aspects of this relationship and the biological significance, as exemplified in this booklet. In the near future, abnormal post-translationally modified proteins will be used as drug targets and diagnostic biomarkers.
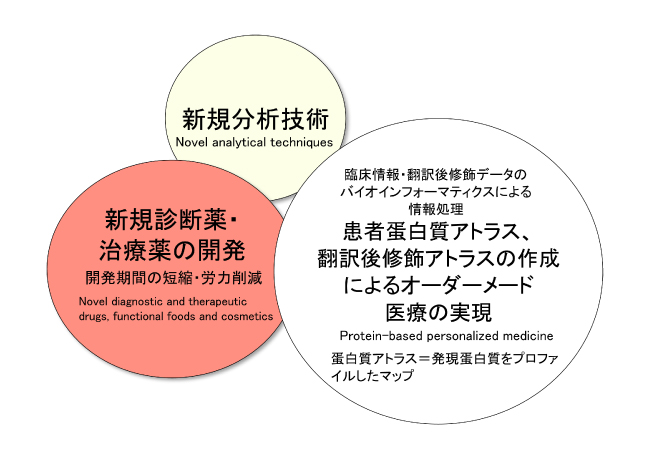
Innovation
In this project, we first expect to develop novel techniques for analyzing post-translational modification, and second, based on these techniques, we plan to facilitate the development of novel diagnostic techniques, therapeutic drugs, functional foods and cosmetics, decreasing the time and energy normally required for development. We expect the construction of a new clinical system using the techniques and drugs developed and the practical application of instruments for diagnosis, diagnostic analysis, and therapeutic drugs. Third, we plan to eventually be able to construct protein post-translational modification maps of individuals (forming a post-translational modification atlas). Using this atlas, we will be able to easily and comprehensively detect aberrant post-translational modifications generated by diseases, offering at this point a novel protein-based personalized medicine, which is strategically different from genomic medicine. These innovations have the potential to catalyze drastic changes in the medical field.