News

| [%article_date_notime_dot%] [%new:New%] |
[%title%] |
|---|---|


Development dev.204239. (2025)
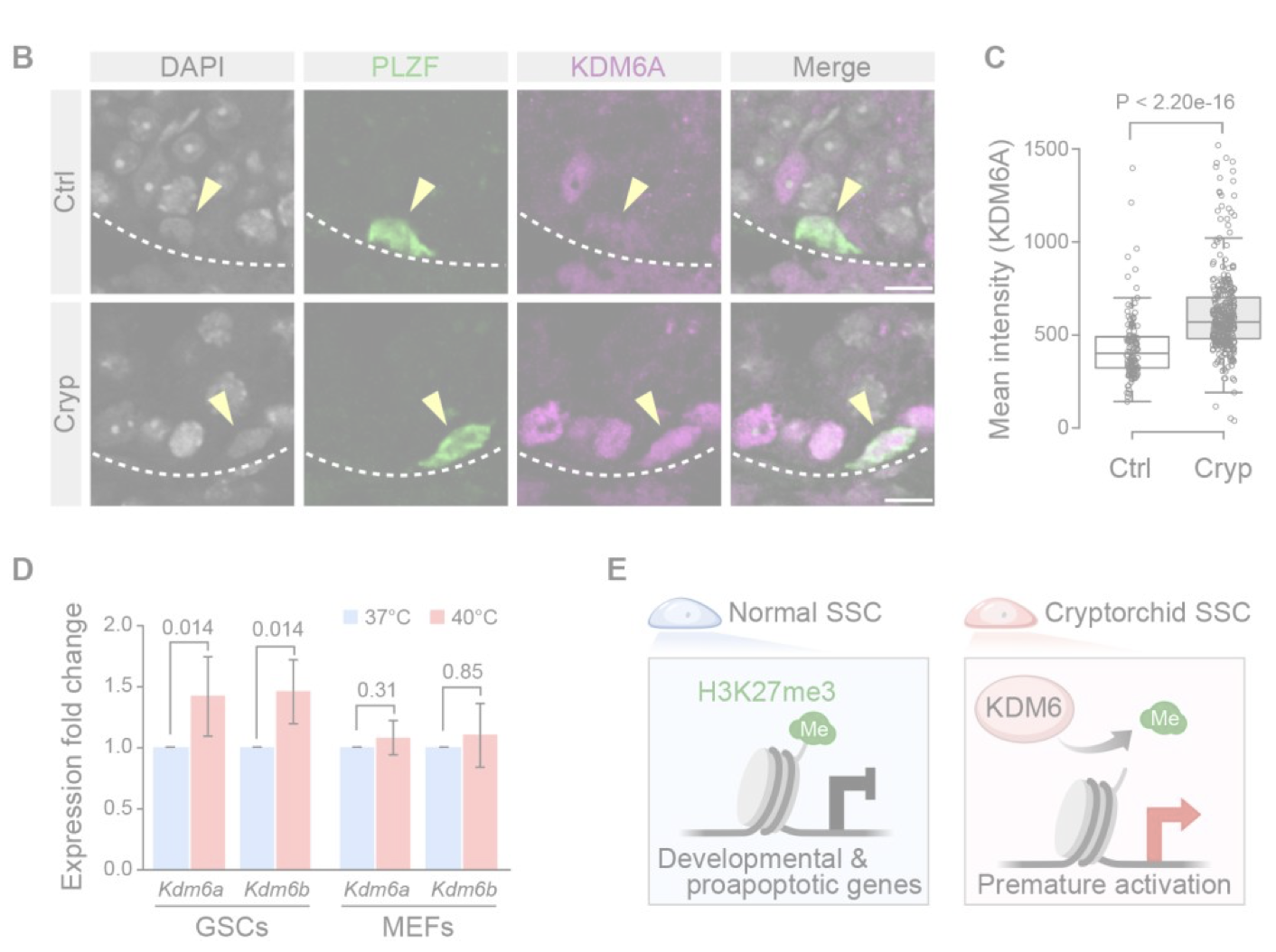
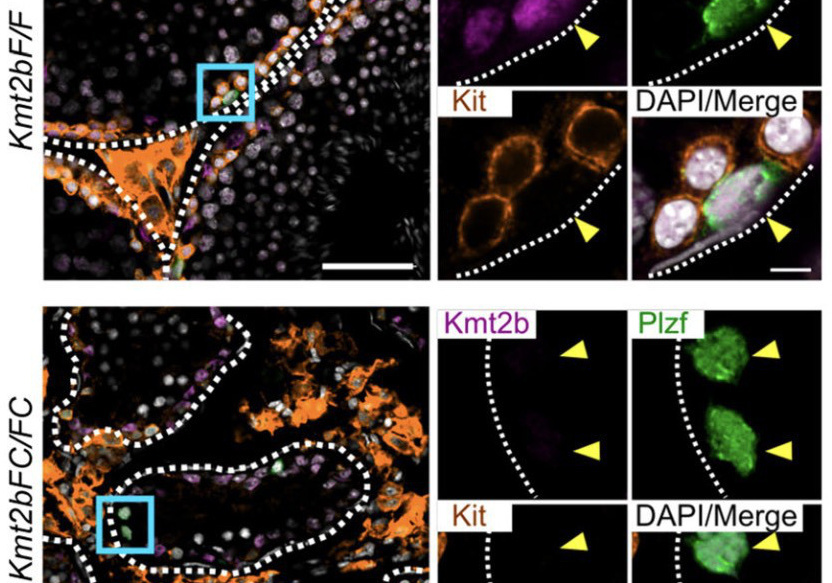
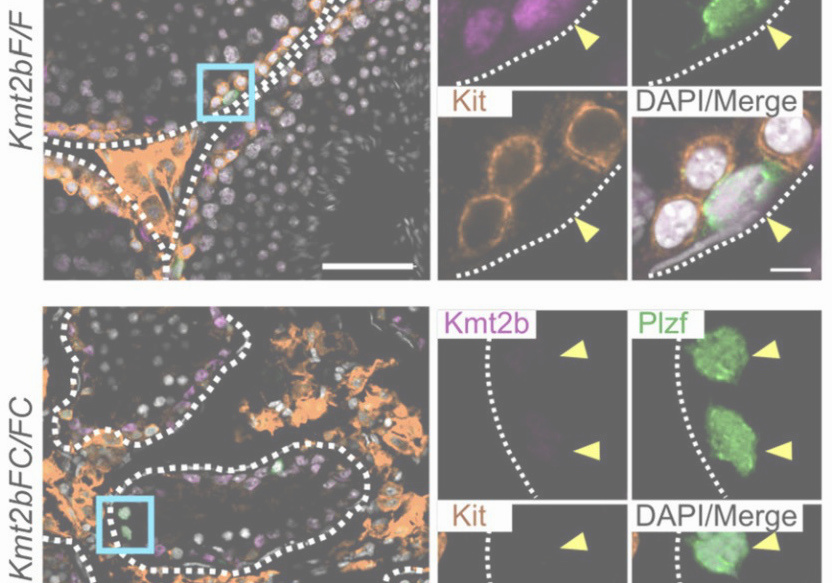
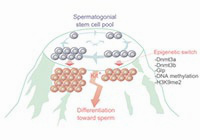
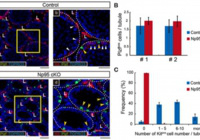
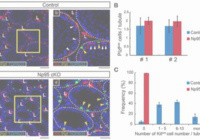
Abnormal H3K27me3 underlies degenerative spermatogonial stem cells in cryptorchid testis.
Kazushige Kuroha, Ivana Dočkal, Uroš Radović, Kuniko Nakajima, Ikue Hoshi, Shion Matsuda, Noriko Kojitani, Kazuyuki Ohbo, Shin-ichi Tomizawa
Gene Expr Patterns (2024) 54, 119383
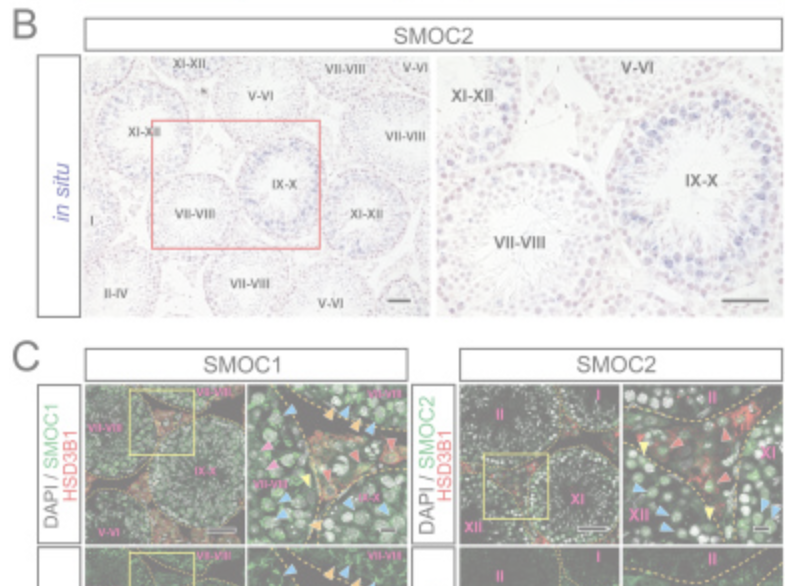
Spatial and temporal expression analysis of BMP signal modifiers, Smoc1 and Smoc2, from postnatal to adult developmental stages in the mouse testis.
M Ono, K Nakajima, S Tomizawa, T Shirakawa, I Okada, H Saitsu, N Matsumoto , K Ohbo
Development (2024) 151(18): dev202834
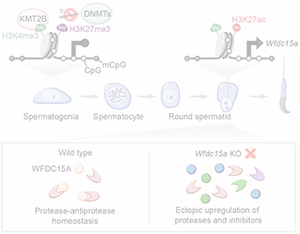
A non-canonical bivalent gene Wfdc15a controls spermatogenic protease and immune homeostasis.
S. Tomizawa, R. Fellows, M. Ono, K.e Kuroha, I. Dockal, Y. Kobayashi, K. Minamizawa, K. Natsume, K. Nakajima, I. Hoshi,S. Matsuda, M. Seki, Y. Suzuki, K. Aoto, H. Saitsu, K. Ohbo
Clinical Epigenetics 13, Article number: 132 (2021)
Oxygen concentration affects de novo DNA methylation and transcription in in vitro cultured oocytes.
Florence Naillat, Heba Saadeh, Joanna Nowacka-Woszuk, Lenka Gahurova, Fatima Santos, Shin-ichi Tomizawa & Gavin Kelsey.
Development (2021) 148, dev194605.
Maintenance DNA methylation in pre-meiotic germ cells regulates meiotic prophase by facilitating homologous chromosome pairing.
Takada Y, Yaman-Deveci R, Shirakawa T, Shari J, Tomizawa S, Miura F, Ito T, Ono M, Nakajima K, Koseki Y, Shiotani F, Ishiguro K, Ohbo K, Koseki H.
Development (2021) 148 (8): dev196212.
Tsga8 is required for spermatid morphogenesis and male fertility in mice.
Kobayashi Y, Tomizawa S, Ono M, Kuroha K, Minamizawa K, Natsume K, Dizdarević S, Dočkal I, Tanaka H, Kawagoe T, Seki M, Suzuki Y, Ogonuki N, Inoue K, Matoba S, Anastassiadis K, Mizuki N, Ogura A, Ohbo K.
Dis Model Mech (2019) 12 (11): dmm040139.
Lack of whey acidic protein four disulphide core (WFDC) 2 protease inhibitor causes neonatal death from respiratory failure in mice.
Nakajima K, Ono M, Radović U, Dizdarević S, Tomizawa SI, Kuroha K, Nagamatsu G, Hoshi I, Matsunaga R, Shirakawa T, Kurosawa T, Miyazaki Y, Seki M, Suzuki Y, Koseki H, Nakamura M, Suda T, Ohbo K.
Development 145 (2018)
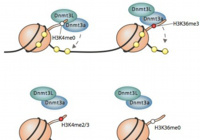
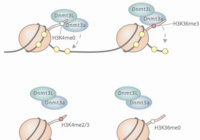
Kmt2b conveys monovalent and bivalent H3K4me3 in spermatogonial stem cells at germline and embryonic promoters.
Tomizawa S, Kobayashi Y, Shirakawa T, Watanabe K, Mizoguchi K, Hoshi I, Nakajima K, Nakabayashi J, Singh S, Dahl A, Alexopoulou D, Seki M, Suzuki Y, Royo H, Peters AHFM, Anastassiadis K, Stewart AF, Ohbo K
Mol Cell Biol. 37(19): e00082-17 (2017)
EPC1/TIP60-Mediated Histone Acetylation Facilitates Spermiogenesis in Mice.
Dong Y, Isono KI, Ohbo K, Endo TA, Ohara O, Maekawa M, Toyama Y, Ito C, Toshimori K, Helin K, Ogonuki N, Inoue K, Ogura A, Yamagata K, Kitabayashi I, Koseki H.
Epigenetics & Chromatin 10: 25-44 (2017)
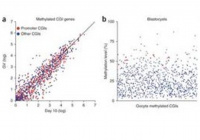
Transcription and chromatin determinants of de novo DNA methylation timing in oocytes
Gahurova L*, Tomizawa S*, Smallwood SA, Stewart-Morgan KR, Saadeh H, Kim J, Andrews S, Chen T, Kelsey G *Contributed equally
Genome Biology 16: 209-215 (2015)
Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape.
Lenka Veselovska, Sebastien A. Smallwood, ... Shin-ichi Tomizawa4, Simon Andrews and Gavin Kelsey*
BioMol Concepts 6(1): 1–9 2015
Epigenetic regulation in stem cell development, cell fate conversion, and reprogramming.
Kazuyuki Ohbo and Shin-ichi Tomizawa
BMC Genomics 16:624-40 2015
DNA methylation and gene expression dynamics during spermatogonial stem cell differentiation in the early postnatal mouse testis.
Naoki Kubo, ……., Takayuki Shirakawa, Hidetoshi Sone, Yasuyuki Sato, Shin-ichi Tomizawa, Kazuyuki Ohbo
Dev Cell., 34: 1-12 2015
The RNA binding protein Nanos2 organizes a post-transcriptional buffering system to retain primitive state of mouse spermatogonial stem cells.
Zhou, Z., Shirakawa, T., Ohbo, K., Sada, A., Wu, Q., Hasegawa, K., Saba, R. and Saga, Y.
Curr. Pathobiol. Rep., 2:1-9 2014
Stem Cell Epigenetics: Insights from Studies on Embryonic, Induced Pluripotent, and Germline Stem Cells.
Shin-ichi Tomizawa, Takayuki Shirakawa, Kazuyuki Ohbo
Development, 140:3565-3576. 2013
An epigenetic checkpoint controls the transition from a stem cell pool to a progenitor cell state in mouse male germ cells.
Shirakawa T., Yaman-Deveci R., Tomizawa S., Kamizato Y., Nakajima K., at.,al.
The International journal of developmental biology 56 867-875 2012
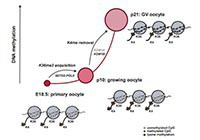
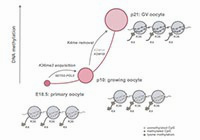
DNA methylation establishment during oocyte growth: mechanisms and significance.
Tomizawa S, Nowacka-Woszuk J, Kelsey G
Nature genetics 43 811-814 2011
Dynamic CpG island methylation landscape in oocytes and preimplantation embryos.
Smallwood SA, Tomizawa S, Krueger F, Ruf N, Carli N, Segonds-Pichon A, Sato S, Hata K, Andrews SR, Kelsey G
Development, 138:4207-4217. 2011
HP1γ links histone methylation marks to meiotic synapsis in mice.
Takada Y., Naruse C., Costa Y., Shirakawa S., Tachibana M., Sharif J., Kezuka-Shiotani F., Kakiuchi D., Masumoto H., Shinkai Y., Ohbo K., Peters A. H.F.M., Turner J. M.A., Asano M. and Koseki K.
Science 332 848-852 2011.
Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus.
Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, Gotoh K, Hiura H, Arima T, Fujiyama A, Sado T, Shibata T, Nakano T, Lin H, Ichiyanagi K, Soloway PD, Sasaki H
Epigenetics & Chromatin 2:5, 2009.
The histone3 lysine 4 methyltransferase, Mll2, is only required briefly in development and spermatogenesis.
Glaser S., Lubitz S., Loveland KL., Ohbo K., Robb L., Schwenk F., Seibler J., Roellig D, Kranz A., Anastassiadis K. and Stewart AF.
Mol Cell Biol. 26:8498-506. 2006.
A CTX family cell adhesion molecule, JAM4, is expressed in stem cell and progenitor cell populations of both male germ cell and hematopoietic cell lineages.
Nagamatsu G, Ohmura M, Mizukami T, Hamaguchi I, Hirabayashi S, Yoshida S, Hata Y, Suda T, and Ohbo K.
Development, 133:1495-1505. 2006.
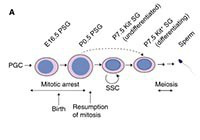
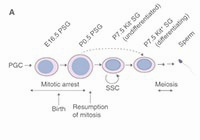
The first round of mouse spermatogenesis lacks the stem cell stage.
Yoshida S, Sukeno, M, Nakagawa T, Ohbo K, Nagamatsu G, Suda T. and Nabeshima Y-I.




大保 和之


富澤 信一


尾野 道男


黒羽 一誠


Kuniko Nakajima


Ikue Hoshi


Aki Hayashi


Noriko Koujitani


Keisuke Minamisawa


Shion Matsuda


Nana Matsuda


Yuga Kashiwagi

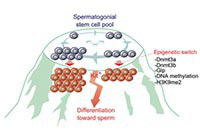
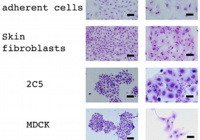
The testicular stem cells have an advantage in that they can be studied with a variety of approaches including a long-term ex-vivo culture, a transplantation assay for evaluation of stemness and histological experiments. We also use other methods such as electron microscopy and super-resolution microscopy to understand their maintenance and differentiation mechanisms.
Research interests
-Analysis of cell fate determination by epigenetic mechanisms
-Study of stem cell proliferation and niche formation
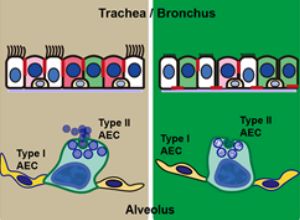
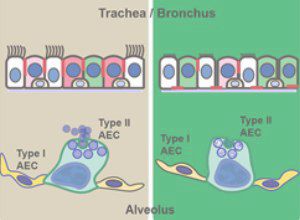
-Mechanistic study of lung diseases associated with alveolar epithelial disturbance
For more information about our graduate programs contact us by e-mail.
E-mail: soshiki@yokohama-cu.ac.jp
Phone: +81-45-787-2567 Fax: +81-45-787-2568
URL: http://www-user.yokohama-cu.ac.jp/~finemorp
Outline
| Department | Histology and Cell Biology, Yokohama City University School of Medicine |
|---|---|
| Address | 3-9 Fukuura, Kanazawaku, Yokohama, Japan |
| Principal Investigator/Professor | Kazuyuki Ohbo, M.D., Ph.D. |
| Study areas | anatomy, histology, epigenetics, germ cells, cell biology |
| Technology | electron microscopy, super-resolution fluorescent microscopy, gene expression and epigenetic analysis (NGS) |
| Member | 4 faculty instructors; 4 technical staff |