April 1, 2024
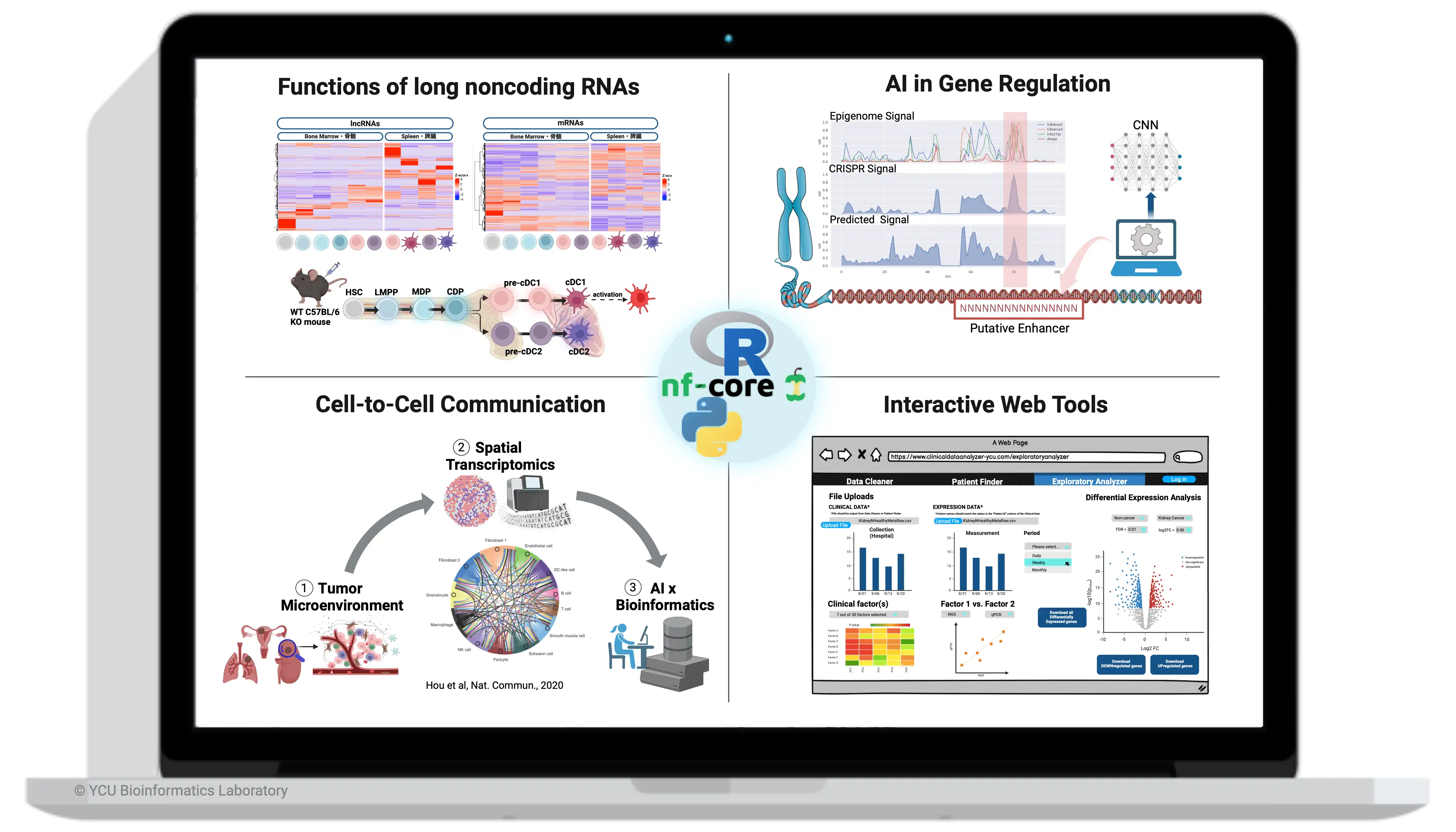
We aim to better understand cell-cell interaction networks in the tumor microenvironment using spatial transcriptomics data and technologies to pave the way for the development of novel cancer treatments.
Introduction
Effective cell-to-cell communication is essential for maintaining body homeostasis, and disruptions in this process often result in serious diseases, including cancer. In 2015 at RIKEN YOKOHAMA, Jordan RAMILOWSKI and Alistair FORREST pioneered a new research field by creating the first draft map of cell-cell communication in human (Ramilowski JA, et al. Nat Commun 6, 7866). After our groundbreaking work, various ligand-receptor prediction tools and databases have been actively developed by us (Hou R, et al., Nat Commun 11, 5011, 2020) and by numerous research groups globally. These research outcomes have been instrumental in investigating diverse biological phenomena and various diseases and in enhancing medical treatments.

Fig: Visualizations of cell-connectivity summary network depict the prediction of cell-cell communication via NATMI (Network Analysis Toolkit for the Multicellular Interactions, Hou et. al., 2020)
Project Specific Aims
- discovery tumor microenvironment ligand-receptor interaction network: cancer cells, healthy cells, and infiltrating immune cells within affected tissues or organs
- enhanced spatial transcriptomic technologies & protocols (in collaboration)
- AI and Molecular Dynamics algorithms for simulating ligand-receptor interactions
- Development of pipelines for Spatial Transcriptomics data analysis
Methods & Data
We employ a combination of experimental techniques (spatial transcriptomics), bioinformatics/AI algorithms, and mathematical models to enhance the accuracy of ligand-receptor predictions. Our approach integrates both publicly available and proprietary Next-Generation Sequencing (NGS) data, encompassing bulk, single-cell, and spatial transcriptomics, as well as proteomics data.
JSPS Postdoc Needed
PhD Researchers with background in (i) Bioinformatics/AI and/or (ii) Biology (basic bioinformatics skills required) are welcome to apply for JSPS Postdoc with us!
Questions? Interested in joining us? Please email Jordan 📧